Today I will be looking at the paper Integrating Graph Contextualized Knowledge into Pre-trained Language Models a biomedical NLP paper about integrating free text and a knowledge graph for entity typing and relation extraction.
In the biomedical domain supervised text training data can be rare while large strutcured knowledge graphs are widely available. Using knowledge graphs to improve BioNLP tasks seemed like an interesting idea.
The paper describes BERT-Medical Knowledge combining
- BioBERT, BERT language model built with open acess Pubmed abstracts and articles
- BERT model for UMLS knowledge graph embeddings initialised with TransE graph embeddings
BERT-Medical Knowledge architecture
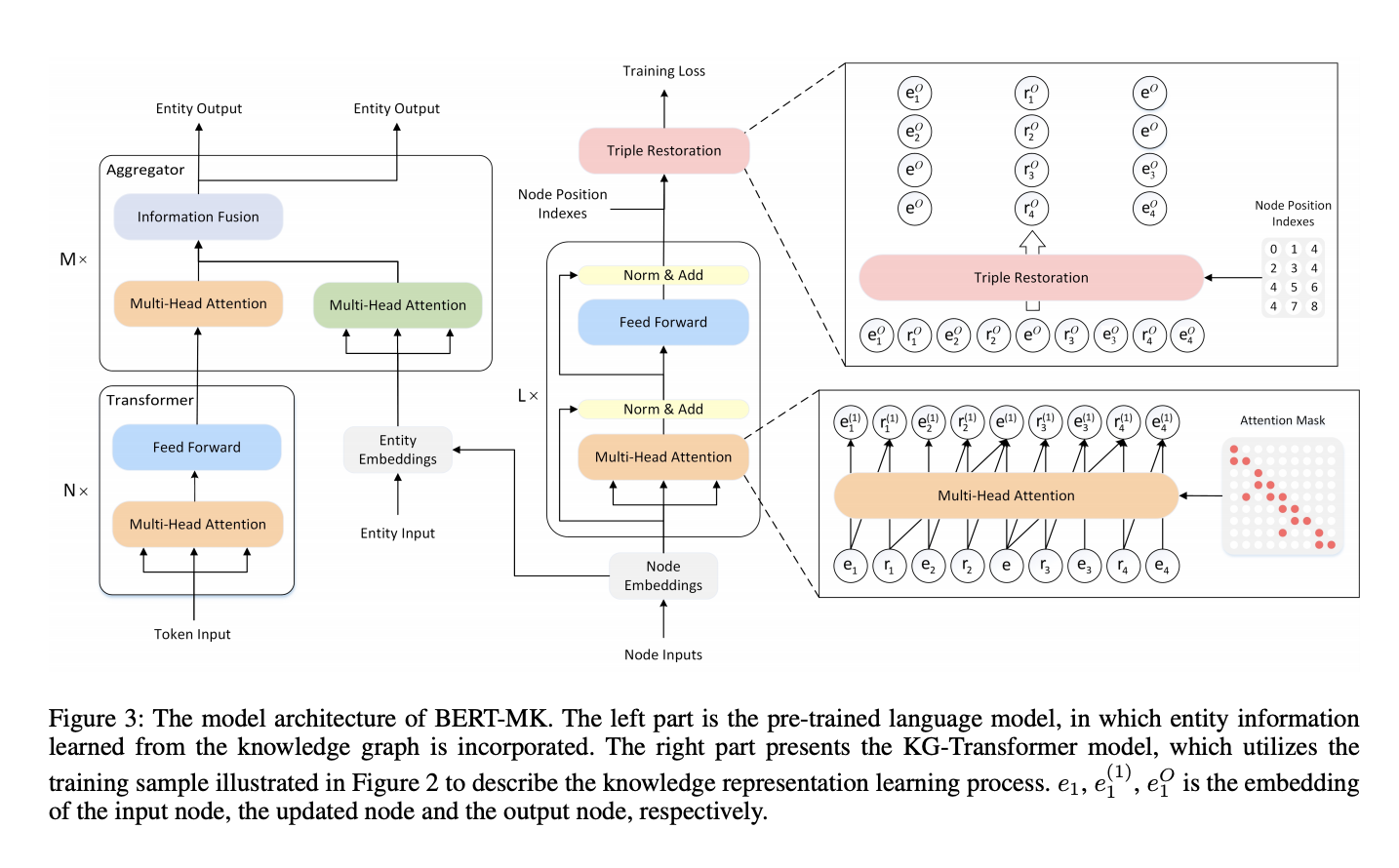
Transformer on the left is BioBERT, Transformer on the right is BERT model for knowledge graph embeddings initialised with TransE graph embeddings. I
How are knowledge graphs used with a Transformer?
- Create a node sequence from an arbitrary subgraph
- Transformer based encoder to encode node sequences
- Reconstruct triples using the translating based scoring function
- Combine node embeddings with Biobert embeddings
###Create a node sequence from an arbitrary subgraph
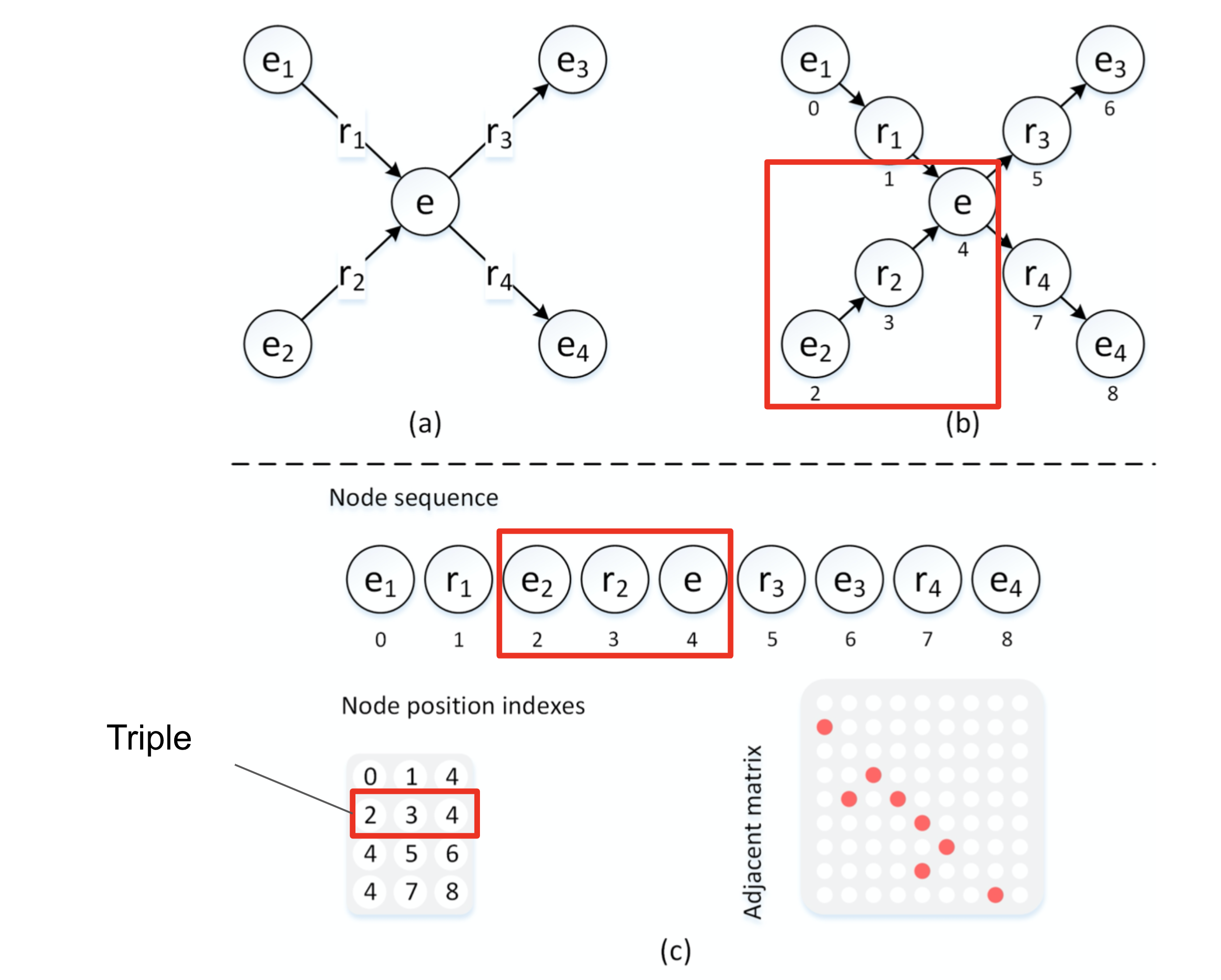
As you can see from the above figure you create a sequence where all nodes that have a directed edge s1 -> r1 -> o1 will be translated into a sequence (s1)(r1)(o1) as you can see relations are jointly learnt as node embeddings. Using the above approach if there are multiple inbound edges then all inbound sequences will be placed before the node it set. So if we also have s2 -> r2 -> o1 then the combined output sequence will be (s1)(r1)(s2)(r2)(o1).
In addition to the node sequences the below are created:
- a node position index P where each row represents a triple
- adjacency matrix A where each value represents and edge between two nodes
Transformer based encoder for node sequences
Seems like a standard Transformer encoder setup with an addition of limiting attention score to in-degree nodes and current nodes.
Concatenate the attention weights
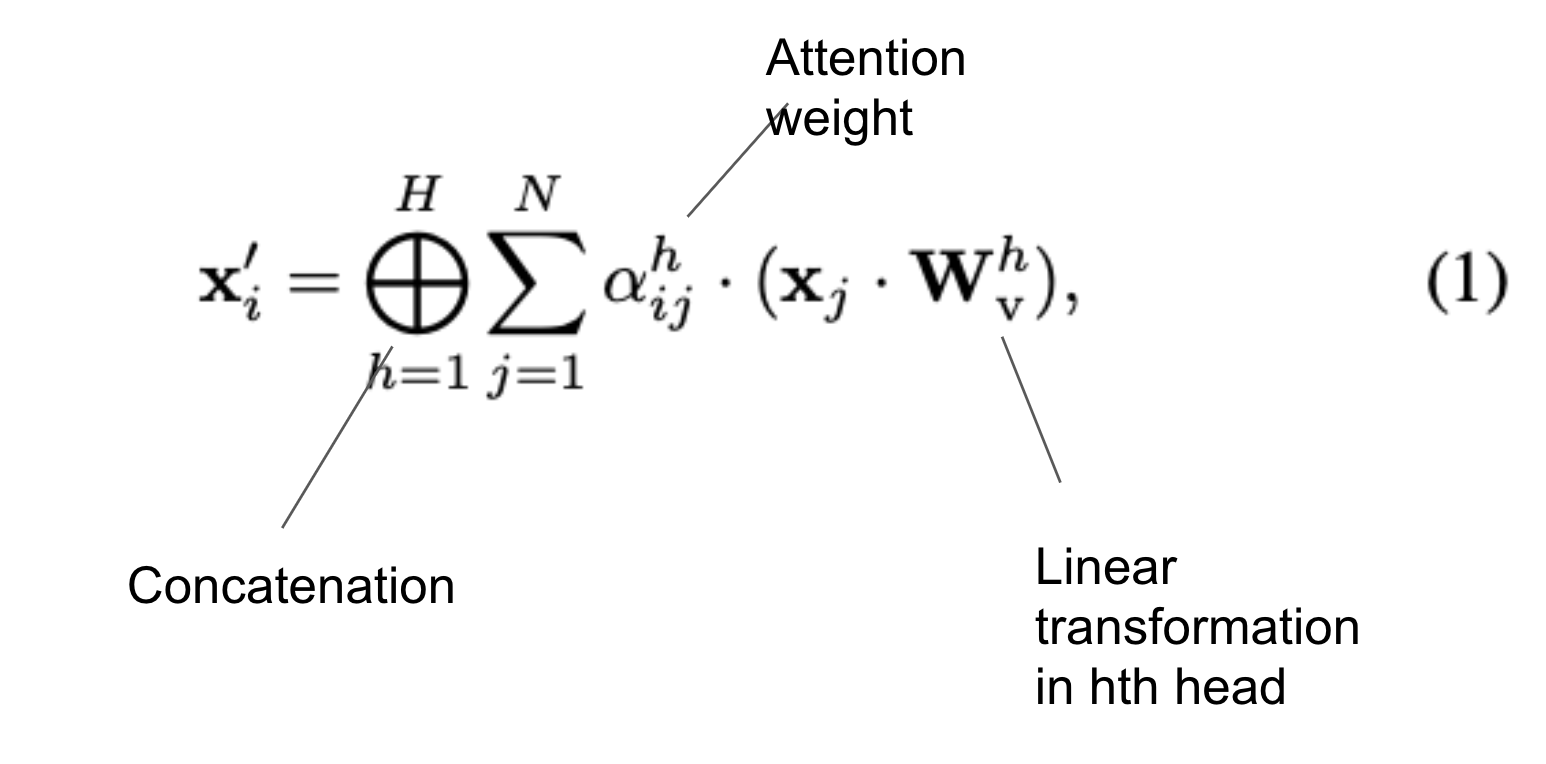
Which are the softmax of the attention score scaled by a constant:
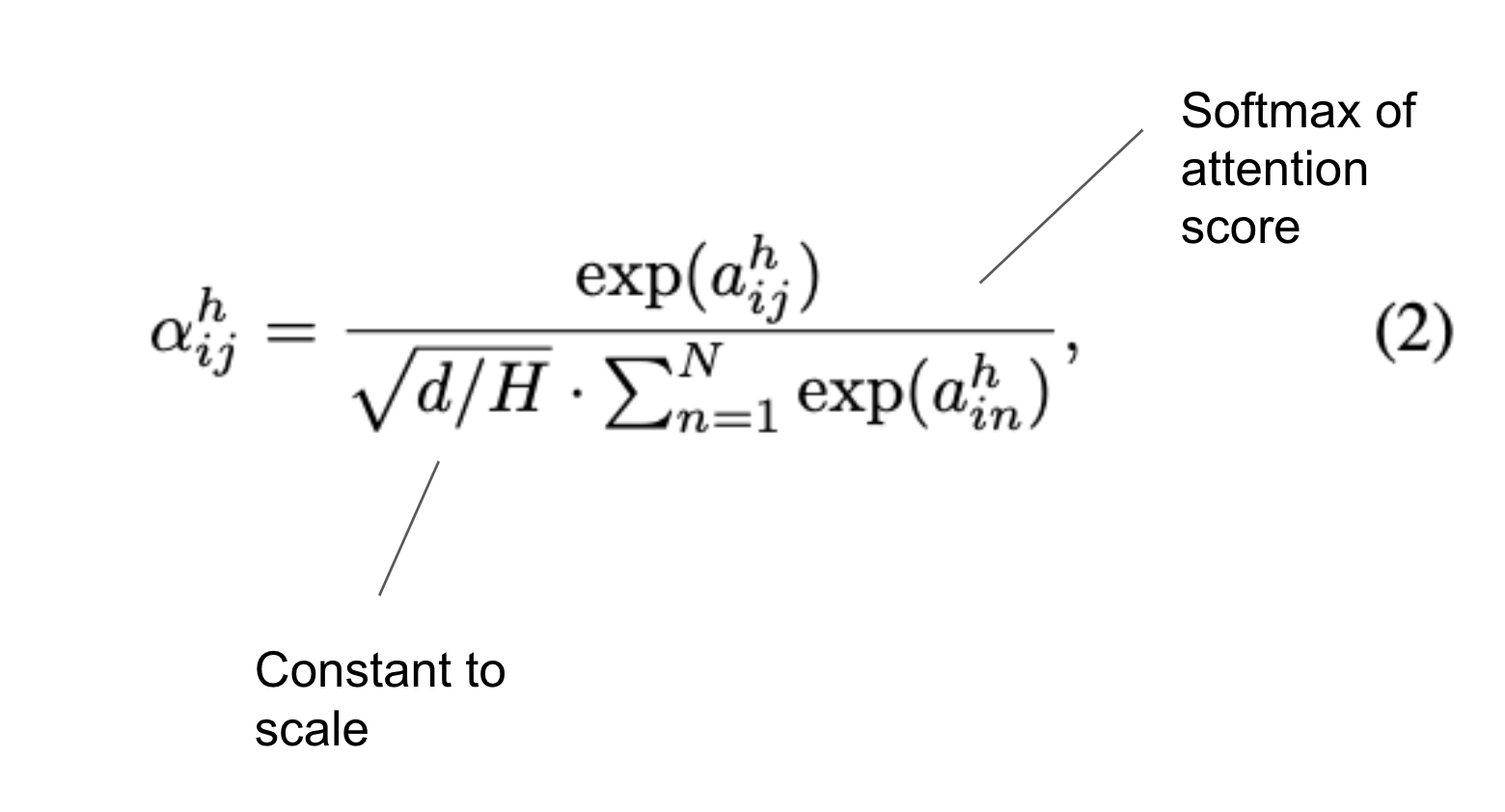
Calculate attention score for degree-in nodes and current node:
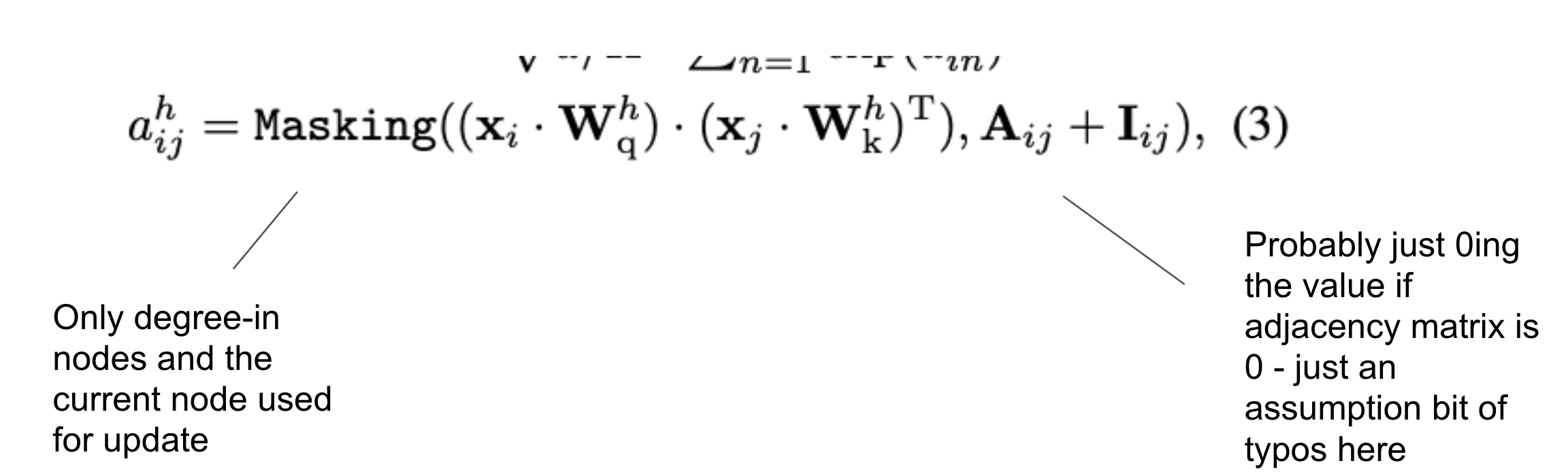
Training objective: Triple Reconstruction
Based on the translation based scoring function reconstruct triples:
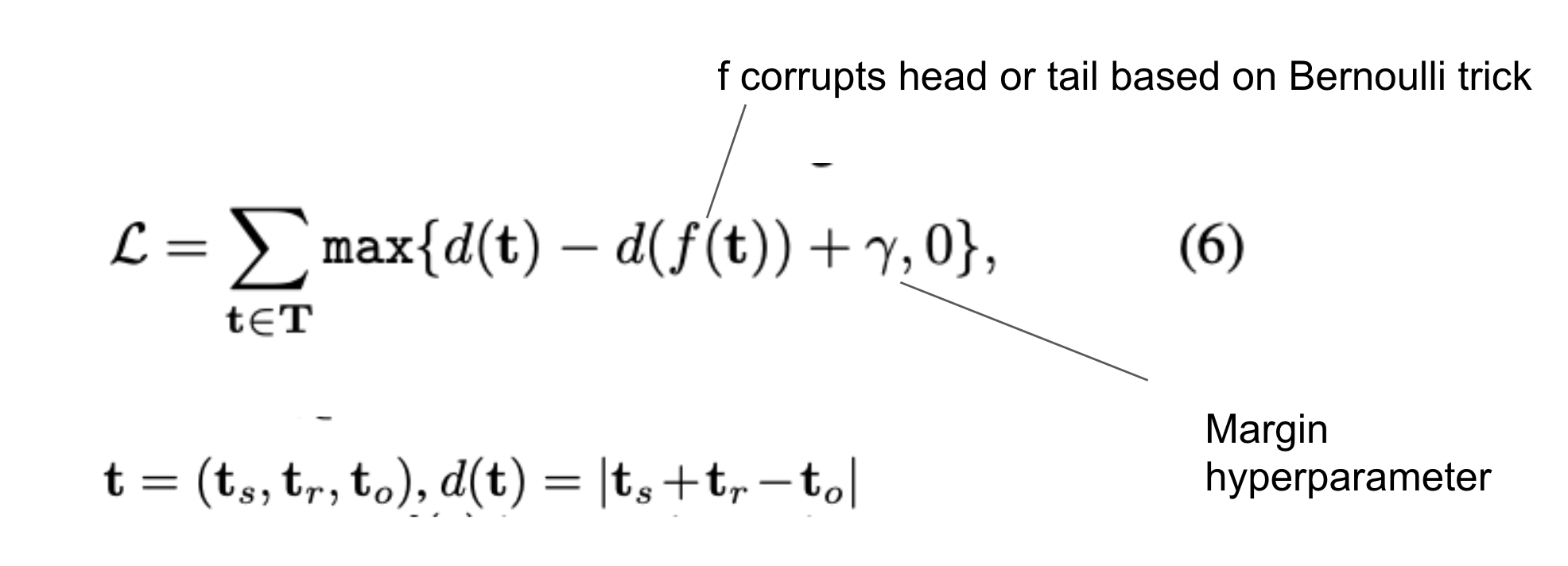
Bernoulli trick
The Bernoulli trick penalises highly connected head or tail nodes so they are more likely to be corrupted. This is based on the assumption that corrupting one of these nodes is more likely to be a true negative rather than corrupting sparsesly connected nodes which could be unknown false negatives.
This is the formula used to decide the probability to corrupt a triple with the Bernoulli trick:
Average number of tails per head - tph
Average number of heads per tail - hpt
Head gets corrupted based on tph / (tph + hpt)
Tail gets corrupted based on hpt / (tph + hpt)
Combining node & BioBERT embeddings
It is not clear from the paper how node and BioBERT embeddings are combined. The architecture diagram just shows an Information Fusion step combining Multi headed attention steps based on the embedding input. Based on this my basic assumption would be the multi headed attention output of the embeddings is concatenated as that would be the most basic form of Information fusion.
Results
Combining BioBERT and ERNIE finetuning the combined model achieves good results as seen below:
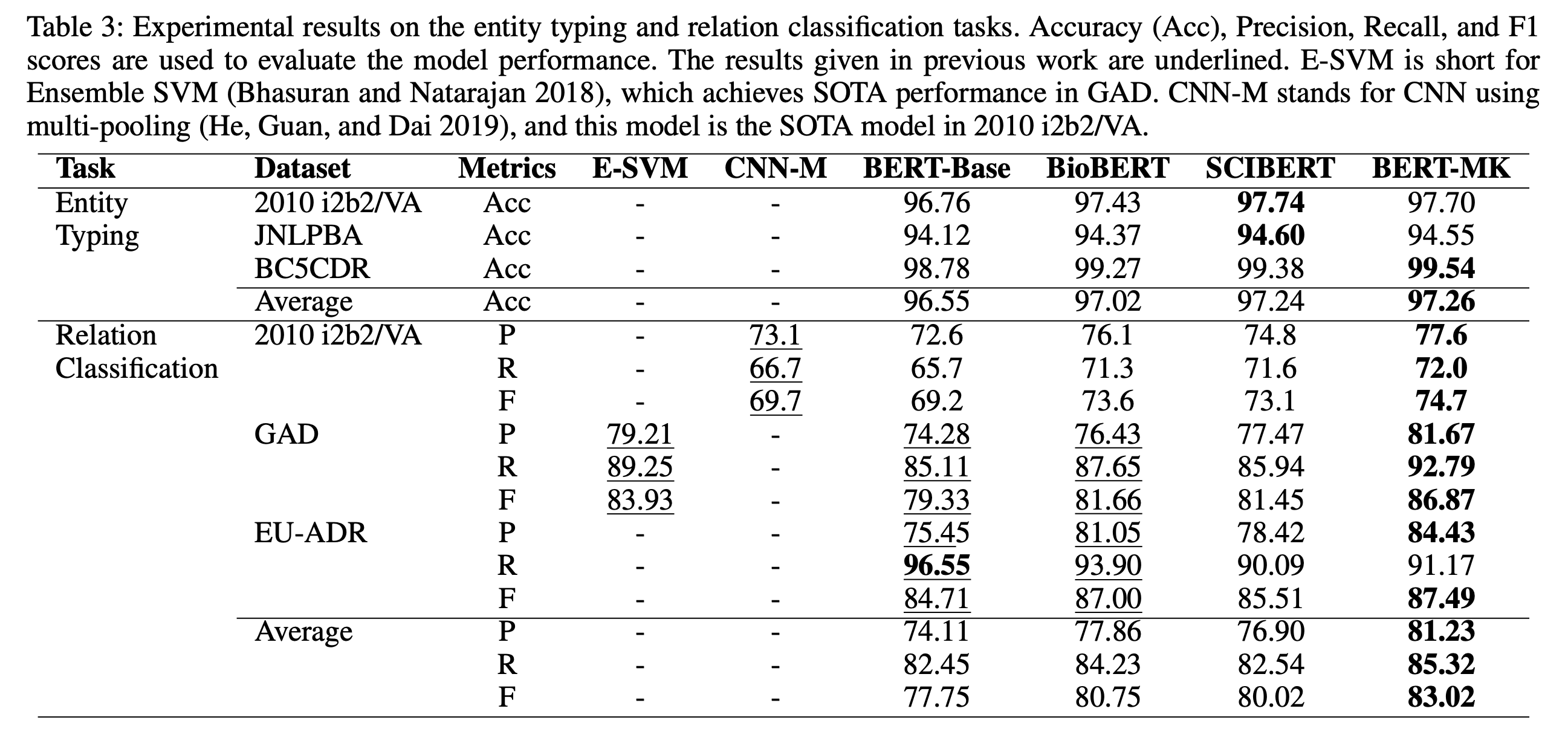
Would be interesting to see this technique applied to other BioNLP tasks in the future. We can surely better leverage the large structured knowledge graphs to improve BioNLP
Credits
All images are from the paper
